Published: Dec 12, 2023 by FME Lab
This time, the 7 partners gathered in Rennes this Tuesday 12th of December and were hosted by STLO at the Institut Agro.

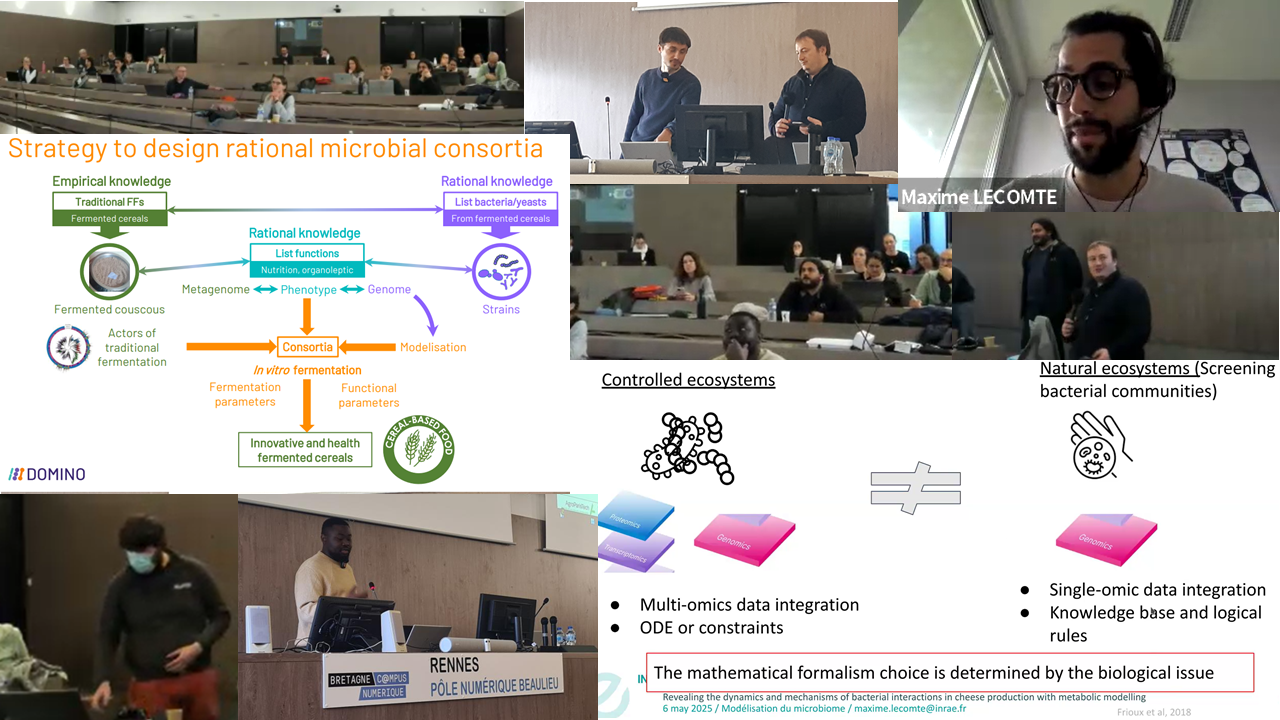
After two years dedicated to prepare the food models for synthetic ecology (Wine and fermented vegetables), the project is now entering into the most exiting (but challenging) part dedicated to the screening of environmental factors and the design of microbial consortia. The project is now ready for integrating many omics data for ecological network modelling. Thanks to Florence Valence for hosting us and for managing the nice diner at crêperie l’épi de blé. We had lively discussions following the presentation of Elham Karimi on genome-scale modelling, minimal community design, key species, and the presentation of Mahendra Maraidassou on fractional factorial design and response surface modelling. The metasimfood project is truly inter-disciplinary, and is taking our teams into a new era of food microbiology.