Food and gut microbiomes science to predict, understand and engineer food microbial communities for health & sustainability.
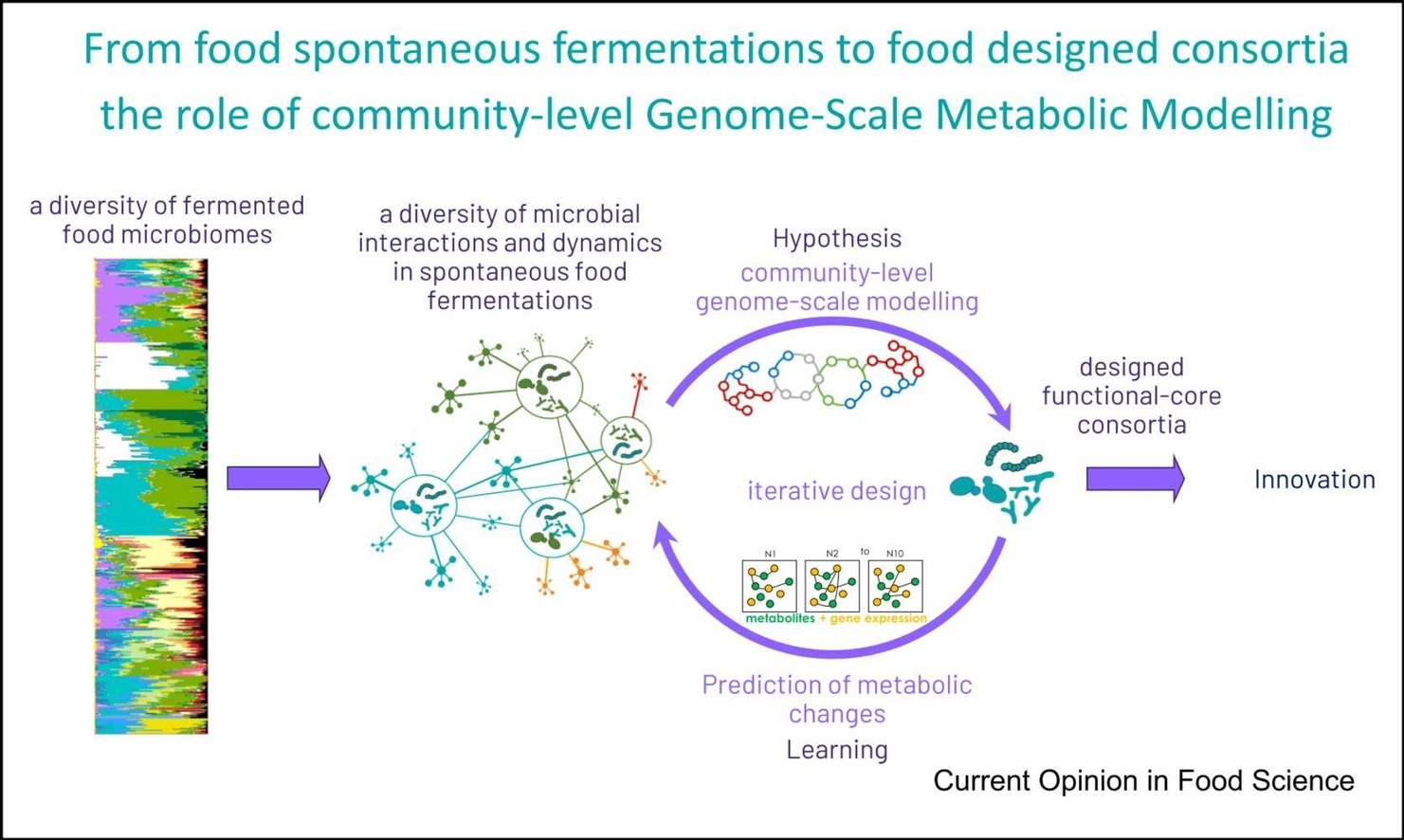
The FME group at MICALIS institute, is focusing its research on food and human gut microbiomes science for the of design food- and digestive-based microbial consortia to advance our understanding of food microbial ecology and how food microbiota interact with human digestive health (functional microbiomes). Our research is focused on the scientific issues associated with community structuring, seeking to determine the impact and role of biodiversity (at different taxonomic, functional and evolutionary scales) in understanding functional population dynamics. As shown in Figure 1, food microbiomes are the cornerstone of health and sustainability issues, in particular in the field of fermented food. Through fermentation of many raw food materials, food microbiomes improve the nutritional impact of many food by enhancing sensory qualities and flavours and by producing vitamins and many other bioactive metabolites having a potential impact on Human digestive health. When ingested as live dietary microbes, food microbiomes can also promote positive interaction with gut microbiomes leading to an equivalent of a domino-like effect for health. Food microbiomes are also promising solutions for the development of new types of plant-based fermented food and for an overall sustainability improvement. However, the versatility of food microbiomes is huge and the idea of harnessing them as a tool for our benefit is limited by the lack of knowledge on how natural communities are shaped and what ecological forces drive their functionality.
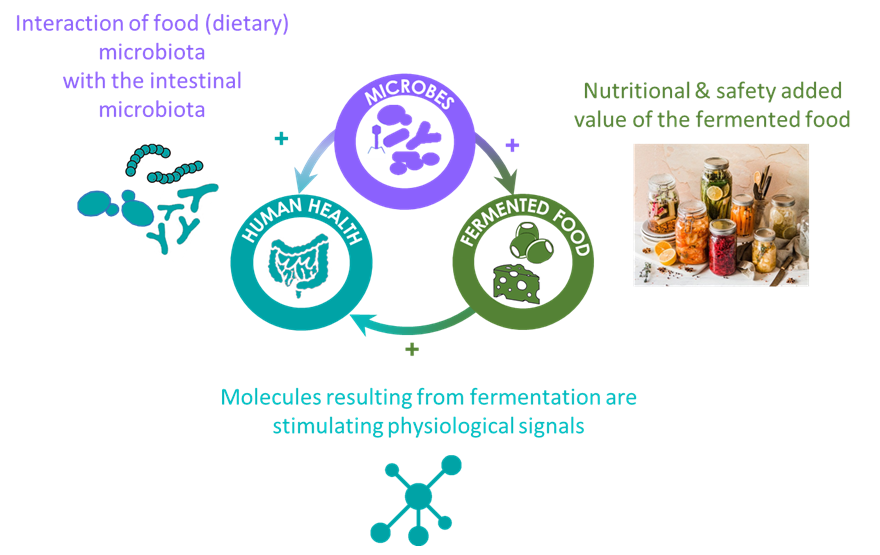
As shown in the Figure 2, our scientific strategy is based on four research axes aiming at a combination of approaches to unlock the immense potential for leveraging food microbial communities as solutions for health and sustainability. A strongly inter-disciplinary strategy combines results and approaches from microbial ecology, high throughput omics data analysis, and computational biology. Our scientific activity creates a strong foreground to develop downstream innovative programs with industrial stakeholders in fermented food areas.
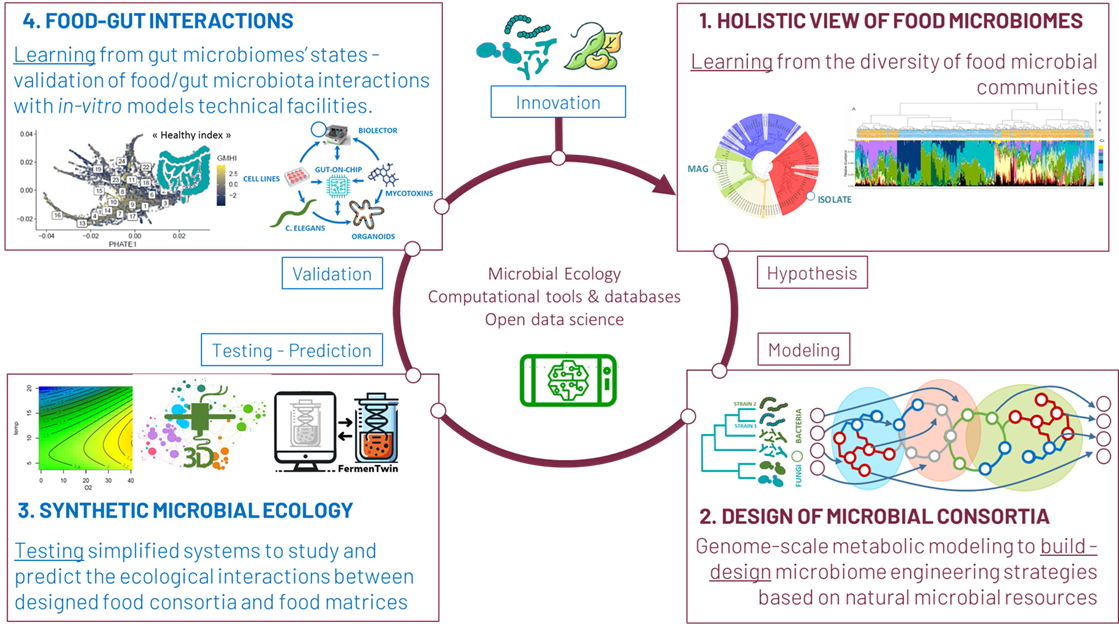
1. Holistic view of food microbiomes in the food chain.
Our first approach is based on a thorough functional analysis of food microbiomes using shotgun metagenomics, comparative genomic and functional analysis of new strains and species isolated from food. Our purpose is to learn from the massive amount of data associated with natural food microbiomes in order to draw hypothesis for the design of functional microbial consortia. In this context, we are active in the development of computational tools and databases on food microbiomes through active collaboration to initiatives in this field at national and European level, in particular within the framework of the EU DOMINO project. Our team has developed a specific focus on an easy-access metatranscriptomic study tool called the F3M tool suite standing for Food Microbiomes Metabolic Modules. Our strategy alternates between integrating metagenomic and gene expression datasets with the production of ecological and technological datasets (see axe 3 below).
2. Design of food microbial consortia.
Our rationale behind the design of new microbial consortia is to rely on the complete knowledge of the genome of individual strains, with which we use genome-scale modelling to assess the overall and idiosyncratic contribution of a strain to a community. In this context, our strategy is to consolidate our biological resources for the design of consortia. Our approach is to structure the constitution of a collection, which need not be very large, but which corresponds to a non-redundant list of species and strains covering the maximum diversity of food microorganisms, in order to serve as a reservoir for the design of consortia built from natural strains. In this axis of our research, we also apply genome- or metagenome-scale metabolic modeling either for food microbiome engineering or the design of consortia. With these methods we leverage genomic data and computational techniques to predict microbial interactions, assesses complementarity among metabolic pathways of different species or strains, and model community dynamics. It enables us the optimization of microbial communities, for example, by constructing minimal communities or identifying key species from microbial populations. We use omics approaches to study the interactions between each member of the community using knock-out/knock-in strategies.
3. Synthetic food microbial ecology.
The simple mapping of populations and of metagenomes is not sufficient to decipher the underlying interactions that operate within microbial communities during their growth in food. Testing the microbial consortia designed in our axis 2 with high throughput facilities and massive data acquisition is also crucial. Our scientific approach focuses on the development of synthetic ecology discipline (knowledge-driven ecological engineering & modelling) applied to microbial communities. We tackle one of the most challenging aspects of food microbial ecology, in particular the role of spatial structuring of communities in solid or semi-solid foods and the influence of specific metabolites on the growth and metabolic activities of microbial communities. Our strategy is based on a partnership and a strong investment in the management of the PIAM experimental platform in the context of the major Ferment du Futur challenge. This project comprises the installation of anaerobic micro-fermenters equipped with microfluidic connectors (BiolectorXT). This equipment, which is an important tool in our synthetic ecology approach, will also be complemented at a later stage (2026, collaboration with team B3D of Micalis) by the acquisition of a 3D printing system for the study and spatialised design of food models. Our “synthetic ecology approach” aims to elaborate hypotheses on the possible metabolic interactions within a microbial community, and to build an experimental approach whose objective is to test them. It is, therefore, a repetitive, iterative strategy whose results are used to reformulate and refine the hypotheses which are then retested experimentally, the aim being always to improve the level of understanding of the interactions when compared to data obtained from real-life food ecosystems.
4. Food-Gut interactions.
Food microbes can have major positive, or negative, impacts on the production of various fermented foods, and in the human gut and digestive health. Although a broad diversity of microbial communities can be identified across many fermented foods, significant information about their contribution to human health is lacking. We aim to understand how diversity of dietary microbes, contribute to a healthy gut microbiome and how the long-term consumption of fermented food contributes to a beneficial health impact. Our objective is to use food and gut microbiome data to allow a more relevant health-orientated design of microbial consortia for fermented food and to identify biomarkers related to the health effect of these fermented foods. For this axis, we collaborate with the French Gut project, in particular on the analysis of the diet-based influence on the different gut microbiomes states. Furthermore, our team is involved in the set-up of in-vitro health models technical facilities for the development of health risk/benefit assessment protocols for food microbial consortia based on food/gut microbiomes interactions.