Published: Nov 6, 2025 by FME Lab
The FME team has published a new preprint in Open Research Europe entitled
“Food Microbiome Metabolic Modules (F3M): a tool suite for functional profiling of food microbiomes.”
Read the article
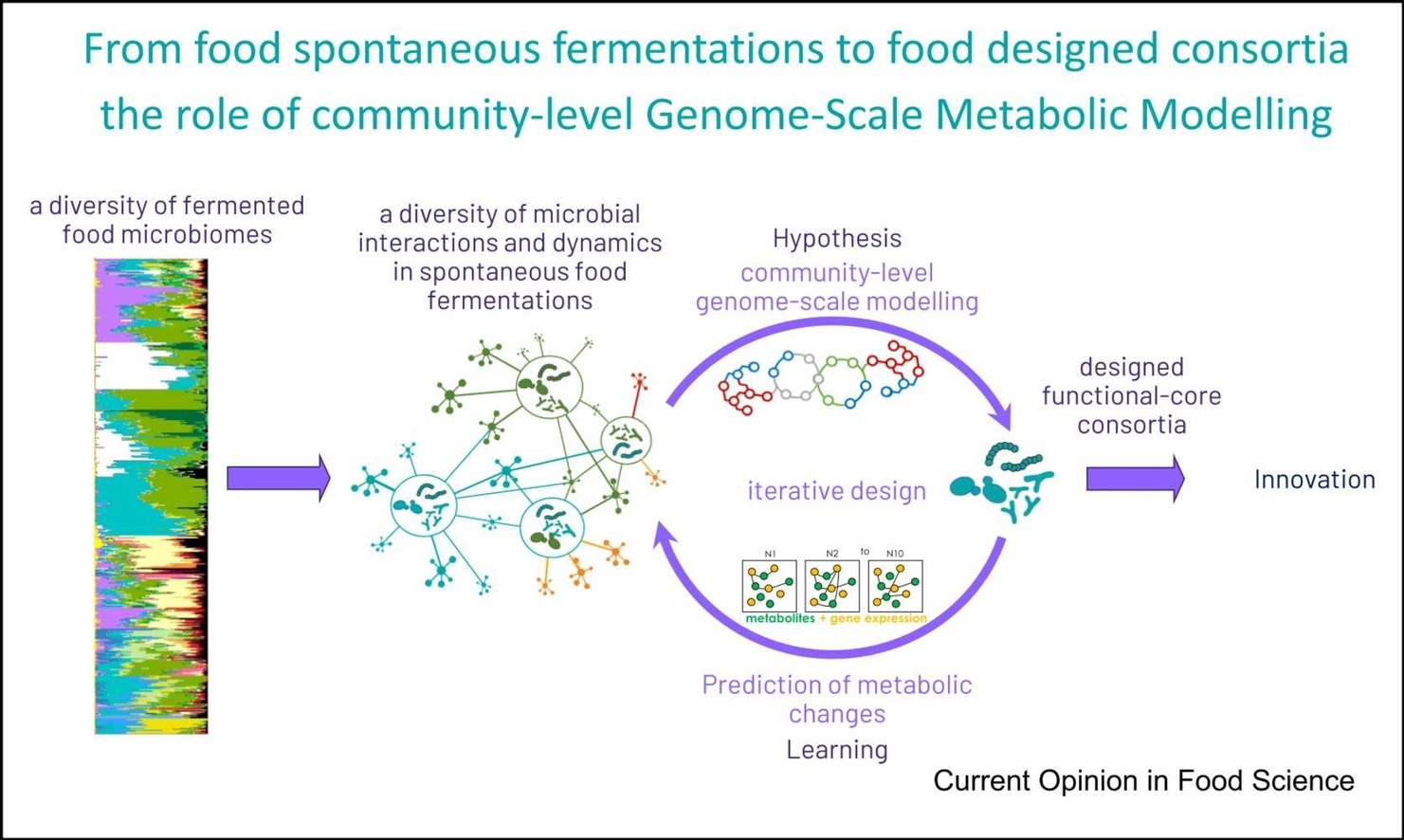
Understanding microbial interactions within food ecosystems is essential for improving the quality, safety, and health properties of fermented foods. However, analyzing these interactions at the functional and metabolic levels remains technically challenging. To address this gap, the FME team developed F3M, an open-source suite designed specifically for the metatranscriptomic analysis of food microbiomes.
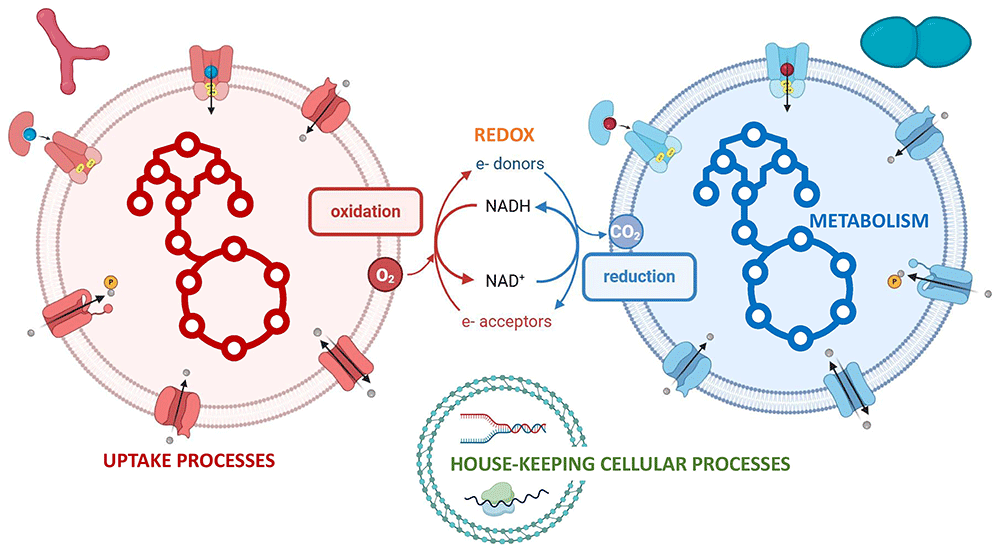
The F3M suite includes:
- A curated database of nearly 2,000 functional genes representing key fermentative reactions
- The F3M Builder, a workflow for constructing annotated gene catalogs and mapping sequencing data
- The f3mr R package, which enables aggregation and analysis of gene expression data across taxonomic and functional levels
Together, these tools provide a coherent framework for exploring metabolic interactions within food microbiomes and for identifying functional signatures associated with fermentation processes.
The article1, authored by Julien Tap, Nacer Mohellibi, Colin Tinsley, Valentin Loux, and Stéphane Chaillou, describes the conceptual framework, design, and open-access resources of F3M.
Access the resources:
Learn and practice with the F3M R tutorial
A complete tutorial and training guide is available online for users who wish to become familiar with the F3M workflow:
https://fme_team.pages-forge.inrae.fr/f3mr/getting-started.html
This hands-on guide introduces the main steps of analysis with the f3mr R package, including:
- Build a reference database from functional and taxonomic annotations.
- Import sample count data .
- Aggregate counts by taxonomic and functional levels.
- Build a count matrix for downstream statistical analyses.
Each section of the tutorial includes example code and data to help users reproduce a full workflow, from raw annotations to interpretable metatranscriptomic profiles.
-
Tap et al. Food Microbiome Metabolic Modules (F3M), a tool suite for functional profiling of food microbiomes. Open Research Europe. 2025 ↩