Published: Oct 1, 2025 by FME Lab
Earlier this month, Julien had the pleasure of participating as a lecturer at the School of Artificial Intelligence Applied to Microbiomes, hosted at AgroParisTech and organized by Aristeidis Litos, Daniel Garza, and Ariane Bize. The event brought together an exceptional group of researchers and students exploring how AI, data science, and mathematical modeling are reshaping our understanding of microbial ecosystems.
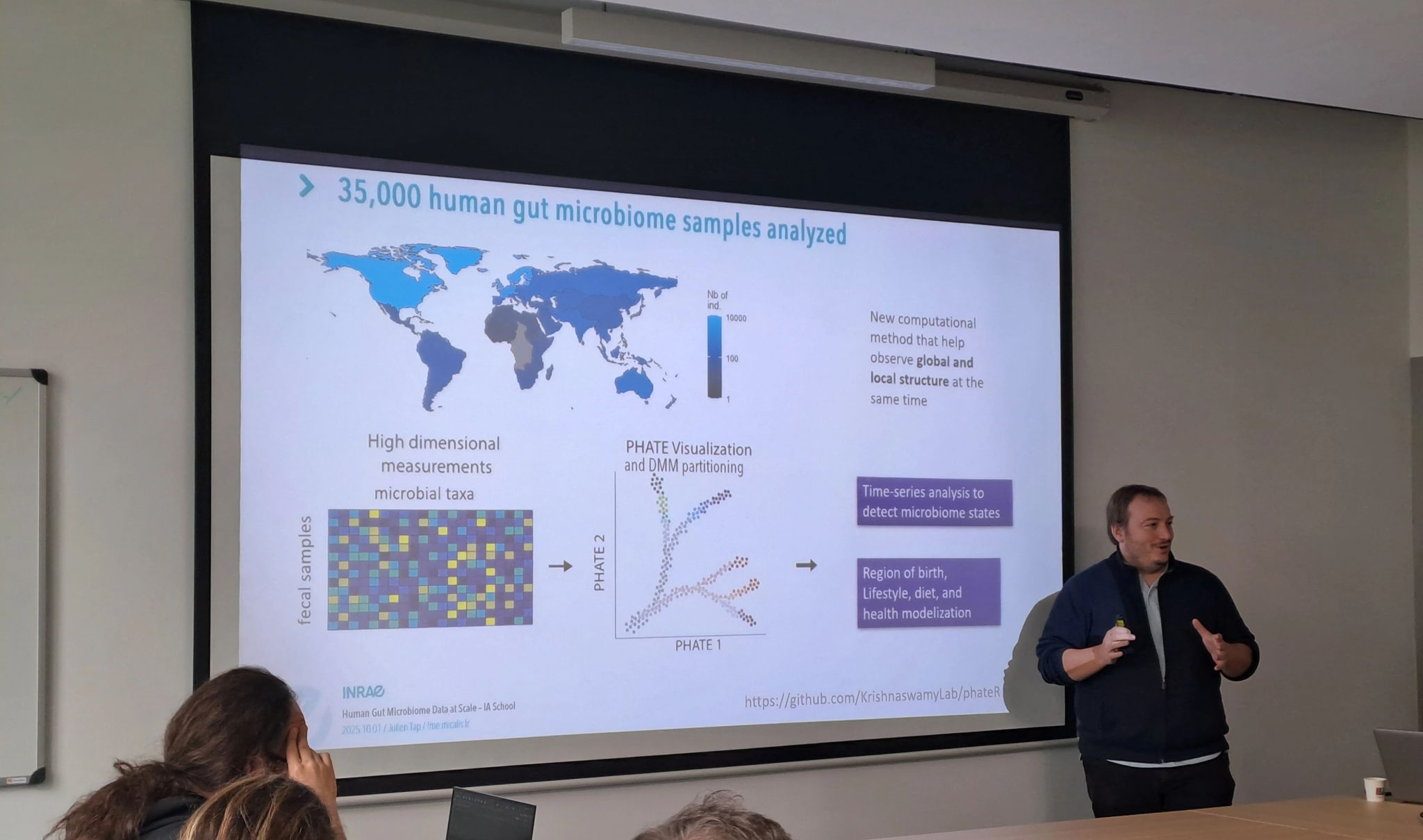
In his lecture1, Julien discussed how large-scale metagenomic datasets—integrating taxonomic, functional, clinical, and nutritional dimensions—can help bridge high-dimensional data with ecological meaning. By examining tens of thousands of gut microbiome samples, we can uncover branching ecological states that describe how lifestyle, diet, and physiology shape microbial diversity. These ecological transitions not only reflect resilience and degradation dynamics but also offer a foundation for precision nutrition and the design of next-generation fermented foods.
A warm thank you to the organizers, speakers, and participants. It’s exciting to see how this field continues to grow, blending theory, computation, and experimentation toward a deeper understanding of microbiomes.
-
Julien Tap. Integrating Human Gut Microbiome Data at Scale: From Individuals to Multi-Layered Features in Clinical and Nutritional Studies. School of Artificial Intelligence Applied to Microbiomes: data analysis, modelling, and engineering of microbiome-based biotechnologies, Université Paris-Saclay, Oct 2025, Palaiseau, France. (hal-05312010) ↩